Did you know that the greatest concentration of bacteria lives in your gut? At two or three years old we have a balanced microbiome. While we know a lot about the human gut microbiome, there is a lot that is unknown about it. There has been a lot of improvement in finding an “unknown microbiome” for example, shotgun metagenomics enables researchers to take a sample of all genes in all organisms and allows them to find an abundance of microbes in many different environments.
What we know: 25 Phyla, ~2,000 Genera, ~5,000 Species, ~80% Metagenome mappability, and 316 million genes
What is unknown?: Undetected unknowns, hidden taxa and strain-level diversity (~20% sequences not matching microbial genomes), functional unknowns (~40% genes without a match in functional databases)
For example, one study where researchers studied a stool sample from 2 lean African men and a stool sample from 1 obese European. In the stool, they found 174 new species never seen in the human gut before and 31 new genome species (which can help in later studies). Found within these new species was, Microvirga Massiliensis which has the largest bacterial genome acquired from a human, along with Senegalvirus which is the largest virus in the human gut. We definitely know a lot more about the human gut microbiome than we did, even though there is a long way to go.
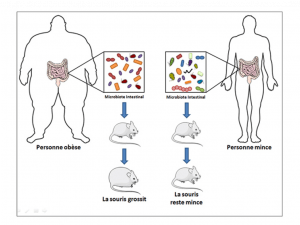
The human gut microbiome intestines in an obese person vs. a lean person
To make improvements in uncovering “hidden strain-level diversity” it is vital to alter sample-specific associations from the metagenomes and to additionally incorporate as many genomes for each species in reference databases. Most species are “open”, meaning they don’t have an upper bound on the size of accessory genomes and it may seem impossible to reclaim all strain-level diversity; however, preserving “the effort of cataloguing strain variants remains crucial for an in-depth understanding of the functional potential of a microbiome.”
The difficulty is that the microbiome contains viruses. The “functional unknown” of the human gut microbiome is the broadest and most challenging to delve and study further into because there is little known about understanding its pathways and genes. There is one creation though, that helped try and find out what was “unknown” about the microbiome, called the Integrated Gene Catalogue. The Integrated Gene Catalogue of the human gut microbiome which consists of 10 million genes. It groups genes into thresholds, thus the genes then fall into sub-units of gene-families. Locating these genes is only a small part of finding out what they actually do. For example, out of 60.4% of the genes that were annotated, 15-20% of the genes have been discovered, but are stilled labelled “function unknown.” These results show how little is known about genes, their functions, and what is current in microbial communities. There is not enough investment in microbiome research. It is difficult because there could be viruses that can be discovered; however, not enough time is being put into finding it.
Lastly, there is a lot of research going into the human gut microbiome. For example, Fecal microbiome transplantation is where stool from a healthy donor gets placed into the other patients intestine, this transplant usually occurs when more bad bacteria take over the good bacteria in the intestine. However, it could cause more disease which is why further investigation in the human gut can solidify that transplantation could overall prevent a bad bacteria take over. The microbiome field is open to all technologies. Understanding the function of the microbiome still remains the largest challenge researchers face, along with the biggest challenge that “targeting specific genes are irreplaceable”, technology should be able to provide solutions (including microbial transcriptome, metabolome, and proteome, and the automation of cultivation-based assays to scale-up the screening of multiple taxa and genes for phenotypes of interest.)


Leave a Reply